I have found out I am actually from West Virginia. Ok, I am just joking.
I knew that my family had a history of marriages among relatives. After all I have only 10 great-great-grandparents instead of the usual 16. With my genome in hand, I set about to quantify the inbreeding.
First, I used David Pike’s Homozygosity tool. It analyzes your genome to find significant runs where the same haplotype is inherited from both parents. Large portions of the human genome are like that. The length of these homozygous regions, however, varies depending on the relation of your parents. If your parents are closely related (first cousins in my case), then you will have longer runs. If your parents are distantly related, then over the generations those genes have had a chance to recombine and so you will have shorter runs that are homozygous.
Overall, the percentage of my autosomal (i.e. on chromosomes 1-22) SNPs that are homozygous is 71.767 and I have 41 runs of homozygosity (ROH) of length at least 200. Here are some of my longest runs:
- Chr 1 has a ROH of length 6009 (30.95 Mb)
- Chr 8 has a ROH of length 5819 (33.00 Mb)
- Chr 9 has a ROH of length 5877 (57.81 Mb)
- Chr 9 has a ROH of length 5941 (24.38 Mb)
Let’s look at my homozygosity percentage by chromosome.
Chr 1: 71.734 %
Chr 2: 69.952 %
Chr 3: 65.741 %
Chr 4: 71.563 %
Chr 5: 69.270 %
Chr 6: 76.025 %
Chr 7: 69.445 %
Chr 8: 72.690 %
Chr 9: 93.323 %
Chr 10: 69.765 %
Chr 11: 71.866 %
Chr 12: 68.443 %
Chr 13: 74.184 %
Chr 14: 68.571 %
Chr 15: 73.087 %
Chr 16: 66.541 %
Chr 17: 77.555 %
Chr 18: 67.763 %
Chr 19: 66.267 %
Chr 20: 66.228 %
Chr 21: 79.902 %
Chr 22: 69.896 %
A majority of chromosomes seem to have reasonable percentages while chromosomes 4, 6, 8, 11, 13, 15, 17 and 21 are high. However, chromosome 9 is really weird: It is 93.323% homozygous.
David Pike writes that:
So far the largest ROHs in 23andMe V2 data that I am aware of consist of:
- 9191 consecutive tested SNPs, corresponding to a DNA segment of length 49.99 Mb.
- 6129 consecutive tested SNPs, corresponding to a DNA segment of length 39.05 Mb.
- 5594 consecutive tested SNPs, corresponding to a DNA segment of length 28.95 Mb.
- 4644 consecutive tested SNPs, corresponding to a DNA segment of length 27.71 Mb.
The highest percentage for overall autosomal homozygosity that I have so far seen from 23andMe V2 data is 71.763%.
As you can see, I am an extreme case.
A number of members at DNA Forums reported their homozygous percentage. Of all those listed, mine is the 2nd highest.
According to the paper Genomic Runs of Homozygosity Record Population History and Consanguinity:
South/Central Asians and West Asians have more than three times as many ROH in all categories over 4 Mb long than sub-Saharan Africans and other Eurasians. 19% of individuals from these populations have ROH over 16 Mb in length, consistent with the high prevalence of consanguineous marriage (marriage between individuals who are second cousins or closer) in these populations.
My total ROH length (segments > 0.5Mb) is about 282Mb which is about 1.2 standard deviations above the Central/South Asian sample mean in that paper. But I am more than 1.7 standard deviations above the mean for longer segments (>5Mb).
Let’s take a look at a graph from the paper’s supplemental material which plots total ROH length versus number of homozygous segments:
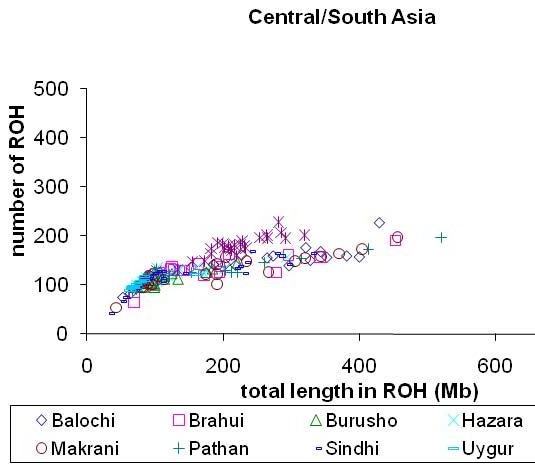
My inbreeding coefficient based on the length of long (>5Mb) runs of homozygosity in my genome (fROH5) is about 0.11 while the average in the Central and South Asian sample for the HGDP dataset is 0.015 (not directly comparable due to different number of SNPs used to calculate).
Finally, I used Plink to calculate my inbreeding coefficient F using all the South Asians from my reference datasets. That coefficient comes out to be 0.1184.
9 is probably the same homolog from your grandmother who comes through both lines, right?
My grandmothers were sisters. So yes chr 9 is likely to have come from there.
Interestingly, the Egyptian ancestry is from the mother of my grandmothers. So the likelihood of chr 9 being completely non-South Asian could be high.
And by high, I mean 50% of course since it’s equally likely to have come from my great-grandfather.
u could run admixture on JUST that chromosome and check 🙂 interesting, i’m more “asian” than my sibling whose results have come back. there should be a lot of variance in people who are recently admixed.
I calculated allele similarity percentage for chr9 with all the reference samples and my chromosome turned out to be too different. 🙂 May be admixture will give better results. Lol
I am also waiting for my sister’s results to compare to mine.
So what would be a “typical” level of homozygosity?
I used the homozygosity tool to analyze 23andme data for me and my wife. My total autosomal % is 70.774%, with 40 runs of 200 or more, longest is 4.88 Mb; hers is 72.339%, with 48 runs of 200 or more, longest is 2.41 Mb.
The typical level varies from one population to another. Those that went through an ancient bottleneck usually have lots of short runs of homozygosity.
Also if you are on 23andme v3, you are likely to get higher percentages than someone on v2.
Recent inbreeding results in you having longer runs. Just like mine does.
If I exclude the no-calls, then the percentage of my autosomal SNPs that are homozygous is 71.392% and I have 147 runs of homozygosity (ROH) of length at least 200. My longest run then becomes 33.49Mb on chromosome 9.
Of course, chromosome 9 stays 92.928% homozygous.
Arriving late to this party. I got only 4 ROHs of over 200 –
Chr 2 has a ROH of length 233 from position 134928799 to position 136509584 (1.58 Mb)
Chr 6 has a ROH of length 270 from position 29689020 to position 30183822 (0.49 Mb)
Chr 6 has a ROH of length 307 from position 57177565 to position 64051004 (6.87 Mb)
Chr 12 has a ROH of length 204 from position 38890224 to position 39114531 (0.22 Mb)
Total autosomal (Chr 1-22): 0.306 % ( 1712 of 560390 SNPs) are NoCalls
Total autosomal (Chr 1-22): 32.058 % (179651 of 560390 SNPs) are Heterozygous
Total autosomal (Chr 1-22): 67.942 % (380739 of 560390 SNPs) are Homozygous (this tally includes NoCalls)
Does this mean I am more mixed than the average South Asian? That would make sense given my family history. How does this compare to the average non-South Asian?
That’s a very low homozygosity percentage for a South Asian.
67.701%
Both parents from the same endogamous caste, and both families have lived in close proximity for many generations within few miles and have connections. It is said everyone in our caste is related to another in the caste and in a few minutes of tracking a connection can be established! But separation was strictly followed for marriage – five degrees on the mother’s side (sa-pinda, of the same body) and unlimited on father’s side (sa-gotra, of the same lineage).