We men inherit the Y chromosome from our fathers who got it from their fathers. So the Y chromosome can be used to trace your paternal lineage. Different sequences of alleles and mutations can be assigned to haplogroups where a haplogroup signifies common descent on the uniparental line.
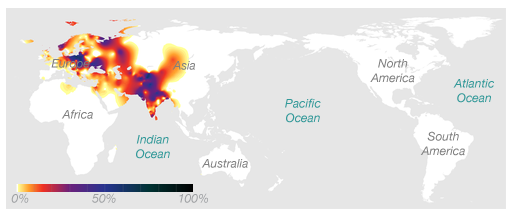
According to my 23andme results, I belong to the paternal haplogroup R1a1a. This group is very common in Eastern Europe as well as South Asia. The distribution of R1a1a can be seen in the map below.
Similarly, we all inherit mitochondrial DNA from our mothers. The sequence of alleles and mutations on the mitochondrial DNA (mtDNA) is also organized into phylogenetic tree.
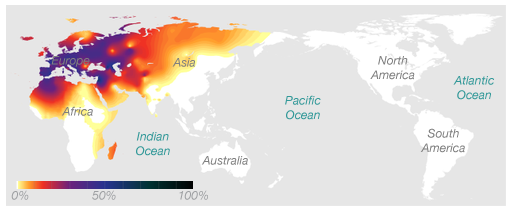
I can trace my maternal line to Egypt (my great-grandmother) and thus I expected a maternal haplogroup common in the eastern Mediterranean. It turns out I belong to haplogroup H, which everyone and their mother belong to in Europe as can be seen in this map.
According to Wikipedia,
Haplogroup H is the most common mtDNA haplogroup in Europe. About one half of Europeans are of mtDNA haplogroup H. The haplogroup is also common in North Africa and the Middle East. The majority of the European populations have an overall haplogroup H frequency of 40%–50%. Frequencies decrease in the southeast of the continent, reaching 20% in the Near East and Caucasus, 17% in Iran, and <10% in the Persian Gulf, Northern India and Central Asia.
Since 23andme didn’t tell me which subgroup of H I belonged to, I used mthap by James Lick:
Your rCRS differences found:
HVR2: 263G
CR: 750G 1438G 4769G 15326G
HVR1: (16519C)Best mtDNA Haplogroup Matches:
1) H
2) H26
2) H(16192)
2) H35
2) H24
2) H10
2) H25
2) H(195)
2) H33
3) H19
Amber’s maternal haplogroup is M4a, which is mainly found in South Asia.
You can see the Y-DNA haplogroup tree and the mtDNA tree online.
My husband has a very similar result:
HVR2: 263G
CR: 750G 1438G 3010A 3796G 4769G 8860G 15326G
HVR1: (16519C)
And he has been put into H1.
By the way…do you know what the parentheses means around 16519C?
According to James Lick: